Vytautas Magnus University (VDU) Research Management System (CRIS) Solution
- 1. Structure of the Research Management System
- 2. DSpace-CRIS
- 3. Research Reporting System (PDB)
- 4. Advantages of the Research Management System
- 5. Research Management Systems in Lithuanian Universities
The goals that are pursued with the help of the VDU research management system:
- To improve the visibility of research output, activities and researchers in the world.
- To accumulate and ensure the long-term preservation of VDU research output and performance results.
- To increase the availability and accessibility of research output through open access.
- To optimize the administration of research activities.
More information about the objectives: Model of accumulation, preservation and dissemination of research output and activities (2018)
Software tools with the help of which the goals are pursued:
| No. | Software product | Purpose | License |
|---|---|---|---|
| 1. | Debian 12 | Debian GNU/Linux is a Linux distribution composed of free and open-source software, developed by the community-supported Debian Project | About licenses found in Debian |
| 2. | Apache Tomcat 10 | DSpace-CRIS backend (REST API) application JEE container (open source) | Apache License, Version 2.0 |
| 3. | Apache Solr 8.11.4 | Indexing and search engine (open source) | Apache License, Version 2.0 |
| 4. | PostgreSQL 15 | Relational database management system (open source) | PostgreSQL License (similar to MIT and BSD) |
| 5. | Node.js 20 | DSpace-CRIS frontend server JavaScript runtime environment (open source) | MIT License |
| 6. | Yarn 1.X | DSpace-CRIS frontend server Angular project JavaScript package manager (open source) | BSD License. About component licenses |
| 7. | Angular 17 | DSpace-CRIS frontend server TypeScript-based, free and open-source single-page web application framework (open source) | MIT License |
| 8. | PM2 | DSpace-CRIS frontend server process manager for the JavaScript runtime Node.js (open source) | AGPLv3 License |
| 9. | Apache2 | Free and open-source cross-platform web server software (open source) | Apache License, Version 2.0 |
| 10. | PHP 8.2 - PHP-FPM | PHP is general-purpose scripting language geared towards web development; PHP-FPM (FastCGI Process Manager) is an alternative FastCGI implementation for PHP (open source) | PHP: PHP License v3.01 (similar to BSD), PHP-FPM: GPL License |
| 11. | JAVA SDK 17 | The Java Development Kit (JDK) is a distribution of Java Technology by Oracle Corporation (open source) | GPL License |
| 12. | Apache FOP 2.9 | Formatting Objects Processor (FOP) is a Java application that converts XSL Formatting Objects (XSL-FO) files to PDF or other printable formats (open source) | Apache License, Version 2.0 |
| 13. | Apache Maven 3.X | Backend server aplication build automation tool (open source) | Apache License, Version 2.0 |
| 14. | Apache Ant 1.X | Backend server aplication runtime build processes automating software tool (open source) | Apache License, Version 2.0 |
| 15. | GeoLite2 | Free IP geolocation database (MaxMind) | GeoLite2 end-user license agreement |
| 16. | Saxon HE 12.7 | XSLT and XQuery processor, free version (Saxon, open source) | MPL-2.0 License |
| 17. | KUSoftas CMS 5 | Content management system, free version (KUSoftas) | License Agreement (Education edition) |
| 18. | DSpace - CRIS 8.2 (2024.02.02) | Free open-source extension of DSpace for the Research Data and Information Management ever developed | BSD 3-Clause License |
1. Structure of the Research Management System

The VDU research management system consists of the following components:
- DSpace-CRIS. This is DSpace 7 with CRIS extension. Metadata and documents of publications, profiles of researchers, research projects and other research items (entities) that can be identified as research production or information related to research are submited here. All this content can be entered using configurable input forms or can be imported from files or external databases, for example: Crossref, Web of Science, Scopus, Pubmed, eLABa, etc. All research entities can be linked to each other (profile of the researcher with the author of the publication, etc.). Indexing and search system ensures the effective finding and presentation of the necessary information.
- Research Reporting System (PDB). This is a WEB information system based on the PHP programming language, Smarty PHP templating system and PostgreSQL RDMBS, with the help of which it is possible to carry out the evaluation of the research production of both the researcher and the department for certification, to compile lists of publications for competitions and other reports on research production. Data for reporting - research publications in CRIS and research projects. Reports can be formed in HTML, PDF, Word, and CSV (Excel) formats.
- University Information System (IS) - a source of information about the University researchers, departments, classification systems, authentication systems, etc. information that is necessary to ensure the functioning of the research management system. As a rule, the information is presented in the form of XML files, but this information can also be provided with the help of the REST API, if there are technical possibilities for this.
- Website. This is a website where the research management system is integrated. In the case of VDU, this is the main website of Unversity https://www.vdu.lt/cris. Such technology ensures more accurate assignment of University research production to the University in external WEB assessment systems, since the URLs of all research entities begin with the URL of the main website of the University: https://www.vdu.lt. On the other hand, both the main website of the University and VDU CRIS focus their efforts and at the same time unanimously raise VDU ratings through the VDU website: both on Google and Google Scholar or other systems.
2. DSpace-CRIS

The system consists of two server software components: Backend and Frontend, which can function both on a single server and on separate servers. The user sends an HTTP Request in the Internet browser of his computer, which is accepted by the CRIS Frontend server. The Frontend server, using the REST API, sends a series of requests to the CRIS Backend server, receives information from there, forms an HTML file and sends it to the user's Web browser as HTTP Response content.
Third-party or University information systems can use all CRIS functionality and data using CRIS Rest API. The system is 100% open architecture, which does not mean that the system is in the public domain. Access rights are fully controlled.
The system provides public access to metadata in the OAI-PMH API, which can be used by external OAI-PMH aggregators, such as: eLABa, OpenAIRE, etc.
More information:
Top2.1 Backend
The DSpace-CRIS Backend is a JAVA EE (Enteprise Edition) application (a functional solution realized in the JAVA programming language), which functions in the Apache Tomcat Java EE container. This is a WEB server that ensures the availability of all Backend functions through the REST API. Authentication is ensured locally or with the help of LDAP (VDU case), Shibboleth, etc. authentication systems. Access to functions and data (research entities, metadata and files) authorization is carried out by combinations of user groups (possible hierarchy) and users. For the authorization of access to data embargo (restriction) until a certain date can be used. After logging in, users automatically acquire the authorization group provided according to the login method, according to which authorization can be performed. The connection from certain IP addresses or IP address zones can also generate additional user authorization groups for the logged-in user, and this can be used to authorize intranet users, for example in VDU network. There is an option to acquire user-assigned groups from LADP.
The DSpace-CRIS Backend uses PostgreSQL RDBMS to store data. The application uses hibernate technology for this, when programs use an object-oriented data form, and the objects themselves are broadcast to SQL data tables and SQL queries in PostgreSQL RDBMS.
The system uses Apache Solr's indexing and search engine. Using the Discovery subsystem, which is based on Apache Solr, it is possible to create indexing and search configurations with great flexibility, create search fields, filter fields, discovery lists, browse lists, etc.
Forms for entering metadata and documents for research entities are fully configured in XML format, and the submission process itself can be managed by setting the workflow steps when the submission takes place, editors receive information about the submitted publication by e-mail. The editor can then assign himself (to edit) the publication, handle it, approve or reject it with a reason. The author receives e-mail messages about the submit process and its results. The submission process can consist of a whole series of forms, they can be authorized to users or groups of users. Form fields can be of various types, it is possible to use key / pair lists for selecting values, controlled dictionaries, authority linked entities, ORCID, etc.
Input forms can be two-level, when an additional form (pop-up) is opened for some input fields and a whole row of fields that are related to the main form field is entered, for example author's affiliation: the name of the department, the code of the department, the country, the part of the deposit, the economic entity. In this way, related metadata can be entered / edited in the form of a table.
At the time of input, it is possible to relate research entities to each other, for example, to associate the author of the publication with the researcher's profile entity, the affiliation with the department entity. In this case, when entering the author's last name, first name or part of the text, a list of profile entities found in the system for selection is automatically provided, where it is possible to select the appropriate profile entity for the linking.
Input forms include the ability to generate Creative Commons (CC) using a dedicated CC license generator.
Authors can edit their own profiles or individual parts of profiles (according to the authorization granted). During the login, the e-mail address of the logged-in user is checked and compared with the e-mail address in the profile. If they match - the user acquires the right to edit his profile. The relation between the login and the profile can also be set by the system administrator manually.
Data can be imported, exported. There is an option for on-line live importing metadata from external systems:
- Pubmed
- ArXiv
- CrossRef
- Scopus
- Web of Science
- SciELO
- VuFind
- OpenAIRE
- European Patent Office (EPO)
- eLABa
- CRIS (import from CRIS itself to ensure copy / paste functionality)
Presentation layout is fully configurable. All tabs, boxes, labels and fields of the represented research entities are configured in a matrix way, using rows and columns for this purpose. Special software renderers are used to represent fields, which are configured and programmable in the Frontend application. All information about rendering is configured in a special Excel file from which it is subsequently installed in the system. Tab and box access authorization for users and user groups is available.
In the system, it is possible to configure various reports of research entities and their lists, formats of citation and export in various ways:
- JSON
- XML
- XML CERIF
- CSV (for lists)
- Excel (for lists)
- XSL (for outputting PDF and RTF (Word) using Apache FOP technology)
- BibTex
- RIS
- Endnote
- CSL citation formats: APA, Chicago, IEEE, ISO, MLA, Harward ir kt.
2.2 Frontend
Frontend Server is a WEB application (similar to Apache Web) that receives and serves users' HTTP requests that they send to the research management system from their web browsers. The Frontend server uses Node.js as a scripting environment (similar to PHP). To program the client's logic, the Angular framework is used by which the prepared programs, along with HTML and CSS, are sent to the user's web browser. There is an opportunity to create your own separate theme of the institution (the way the interface is represented). The Frontend server communicates with the Backend server via the REST API by realizing the functionality of the research management system and presenting it to the user.
Frontend has its own cache mechanism. There is a BOT cache (for Internet indexing systems, e.g. Google) and Anonymous cache (for unregistered users).
The system has message / translation files in English and Lithuanian (technically, the amount of languages is not limited).
The amount of frontend processes can be controlled with the help of a PM2 monitor. Each process can use a separate virtual processor.
Top2.3 REST API
All DSpace-CRIS functionality is realized and accessible through the REST API. The data exchange format is JSON. Access to the REST API is: https://portalcris.vdu.lt/server. For authorization, Bearer tokens are used, which are received after the login (sent as the Authorization Header) or such a token can be created in the CRIS user profile area. The use of the REST API may be restricted by IP addresses.
REST API uses HTTP Request Methods:
- GET. Used to read an object
- POST. Used to create an object
- PUT. Used to overwrite the contents of an object
- PATCH. Used for replacement / removal of parts of the object
- DELETE. Used to remove an object
PATCH Method makes it possible to control the content of an object in parts:
- Remove metadata
- Add metadata
- Replace metadata
- Change order of metadata
REST API example: get entity item 20.500.12259/57984 metadata: https://portalcris.vdu.lt/server/api/core/items/5d9ace22-691c-4051-bbb4-d918748c5838.
Top2.4 OAI-PMH
CRIS OAI-PMH is available in the URL https://portalcris.vdu.lt/server/oai. It consists of the following contexts:
- Default Context (base address: https://portalcris.vdu.lt/server/oai/request). This context is to be used in the case of non-specialized, general-purpose OAI-PMH aggregators. Supported metadata formats: uketd_dc, qdc, didl, mods, ore, mets, oai_dc, rdf, marc, dim (vidinis sistemos formatas), etdms.
- OpenAIRE Data Repository Context (base address: https://portalcris.vdu.lt/server/oai/openairedata). This context is to be used in the case of OpenAIRE in accordance with the Rules of the Data Archive OpenAIRE. Supported metadata formats: oai_dc, oai_datacite.
- OpenAIRE Context for CRIS System (base address: https://portalcris.vdu.lt/server/oai/openairecris). This context is to be used in the case of OpenAIRE according to the rules of the OpenAIRE CRIS system v 1.1.1. Supported metadata formats: oai_dc, oai_cerif_openaire.
- Driver Context(base address: https://portalcris.vdu.lt/server/oai/driver). This context is to be used in the case of driver. Palaikomi metaduomenų formatai: didl, mets, oai_dc.
- OpenAIRE Context (base address: https://portalcris.vdu.lt/server/oai/openaire). This context is to be used in the case of OpenAIRE in accordance with the rules of the OpenAIRE Guidelines for Literature Repositories v3.0. Supported metadata formats: mets, oai_dc.
- OpenAIRE 4 Context (base address: https://portalcris.vdu.lt/server/oai/openaire4). This context is to be used in the case of OpenAIRE in accordance with the rules of the OpenAIRE Guidelines for Literature Repositories v4.0. Supported metadata formats: oai_dc, oai_openaire.
Converting entity metadata to OAI-PMH is controlled by XSL transformations and can be edited. The structure of the record identifiers is as follows: oai:vdu.lt:handle, where handle is the handle of the entity. For example: oai:vdu.lt:20.500.12259/57984. OAI sets, as a rule, correspond to CRIS communities and collections. Standard structure of identifiers for OAI sets: com|col_handle prefix_handle number. For example: The OAI set ID of the PDB collection (20.500.12259/36910) is col_20.500.12259_36910. There may also be specialized sets, for example: openaire. Examples of OAI-PMH queries include:
- Information about the OAI-PMH server: https://portalcris.vdu.lt/server/oai/request?verb=Identify
- List of sets: https://portalcris.vdu.lt/server/oai/request?verb=ListSets
- List of metadata formats: https://portalcris.vdu.lt/server/oai/request?verb=ListMetadataFormats
- PDB collection entity indentifiers: https://portalcris.vdu.lt/server/oai/request?verb=ListIdentifiers&metadataPrefix=oai_dc&set=col_20.500.12259_36910
- PDB collection entity metadata in oai_dc format: https://portalcris.vdu.lt/server/oai/request?verb=ListRecords&metadataPrefix=oai_dc&set=col_20.500.12259_36910
- Entity metadata in DIM format: https://portalcris.vdu.lt/server/oai/request?verb=GetRecord&metadataPrefix=dim&identifier=oai:vdu.lt:20.500.12259/57984
More information about OAI-PMH: The Open Archives Initiative Protocol for Metadata Harvesting.
Top2.5. Functionality
DSpace–CRIS functionality:
- Indexing and search (Discovery):
- Indexing, search.
- Filters and facets.
- Top lists.
- Browse lists
- Presentation layout:
- Matrix method: rows, columns.
- Tabs, boxes, metadata, renderers, authorization.
- Modeling in an Excel environment and loading into the system.
- Authentication: IP, Password, LDAP, SSO, ORCID, etc.
- Authorization: groups, users, embargo, policy.
- Configurable submission:
- Steps.
- Forms.
- Workflow.
- Working area for each registered user (workspace).
- Importing metadata from external live systems: eLABa, WOS, SCOPUS, CROSSREF, Pubmed, etc.
- REST API, OAI-PMH.
- Etc.
CRIS data structure items:
- Community – a set, can have other sets in it.
- Collection – collection is an integral part of a set.
- Entity – (object) - component of a collection::
- Person – researcher's profile.
- Project – research project.
- Funding – source of funding.
- OrgUnit – department.
- Journal – journal.
- Publication – publication.
- ETD – thesis / dissertation.
- Product – research data.
- Standard – standard.
- Patent – patent.
- Event – event.
- Equipment – research equipment.
- Database – the database / publisher that refers the publication.
- List – lists of approved databases / publishers, which are used in research evaluation.
3. Research Reporting System (PDB)

3.1. Reporting Request Engine
Research publications are prepared in advance for their subsequent use in reports, supplementing them with additional attributes necessary for accounting, for example: WOS IF, WOS AIF, Scopus SNIP, etc. Additional classifications necessary for reporting (this is configurable) can be created, for example: sections of the report, which are compiled according to science fields, type of publication, WOS and SCOPUS indicator, referencing DB, approved publishers, etc. All these data are prepared in advance, and their structure is known in advance. Therefore, when creating reporting templates, you do not need to know the internal structures of the database, know the technologies of DB queries, etc.
Additional attributes of publications are created in the following processes:
- An publication entity was created in the DSpace-CRIS system, a correction was made to the entity, the entity was removed. In these cases, the DSpace-CRIS system with the help of the REST API automatically contacts the research reporting system and changes are made to the corresponding record in the research reporting system.
- The administrator initiates an analogous REST API request to a entity based on its handle. The metadata of the entity are transmitted from DSpace-CRIS to the research reporting system (or removed if it is a request from the removal REST API) and changes are made to the corresponding record in relation to the research reporting system.
- The administrator initiates the preparation of all records (all publication metadata from DSpace-CRIS to the research reporting system can be re-transferred beforehand).
Additional classifications for reporting, e.g. the codes of the sections of the research production evaluation report (1.1, 1.2, ..., 3.1, 3.2, etc.) can be developed additionally (in addition to the already predetermined ones, such as WOS indicators, Scopus indicators, afield data, etc.). In this case, for each such classification, XSL files are created, which, by means of the XSLT transformation, create the necessary section code and assign it to publication. In addition, an additional classifier is created for this classification, with the help of which it is possible to find out the name of the section in Lithuanian and English according to the section code and subsequently use it in the reports. Parameters such as: WOS indicators, Scopus indicators, type of publication, research fields, author's araffiliation data, belonging to the approved lists of databases and publishers, etc., are transmitted to the XSL file as parameters. At the same time, all the metadata of the publication in DIM format (for example: entity 20.500.12259/57984 via OAI-PMH DIM format), are provided, which can be additionally used to determine the classification code with the help of XSL XPATH expressions.
The reporting request engine, upon receipt of a request with certain filtering attributes, selects research publications and returns all their data to the report in the list in JSON format.
Top3.2. Reporting Smarty Templates and Plugins
Reports Smarty templates use the PHP Smarty template engine. Smarty's purpose is to separate presentation (HTML/CSS) logic from application server (PHP) logic. The template engine allows you to create templates using Smarty variables, modifiers, functions and comments. Templates can have hierarchical structures where a lower-level template inherits properties and variables from a higher-level template. Templates can simply be included to each other. Smarty has logic control blocks: "for", "foreach", "if" and etc. New blocks and functions can be created using the PHP programming language, and PHP sentences can be used in the templates themselves.
Smarty plugins in the PHP programming language are created for Smarty templates integration with the REST API of the reporting request engine:
- The purpose of this plugin is to take over the main parameters of the request, which are generated in the user interface environment, to refine them, to pass them to the query engine, and after receiving the answer - to place the results in the Smarty variable in the publication list form. In the Smarty template, this list can be iterated, the data of each publication is processed and inserted into HTML code.
- Project plugin. The purpose of this plugin is to select projects in which the researcher is involved in the preparation of the report.
- Classifier plugin. The purpose of this plugin is to determine the text of the name by the classification code scanned from the publication data in the list of results.
- Other plugins, which are designed to ensure the functionality of the system.
3.3. User Interface and System Implementation
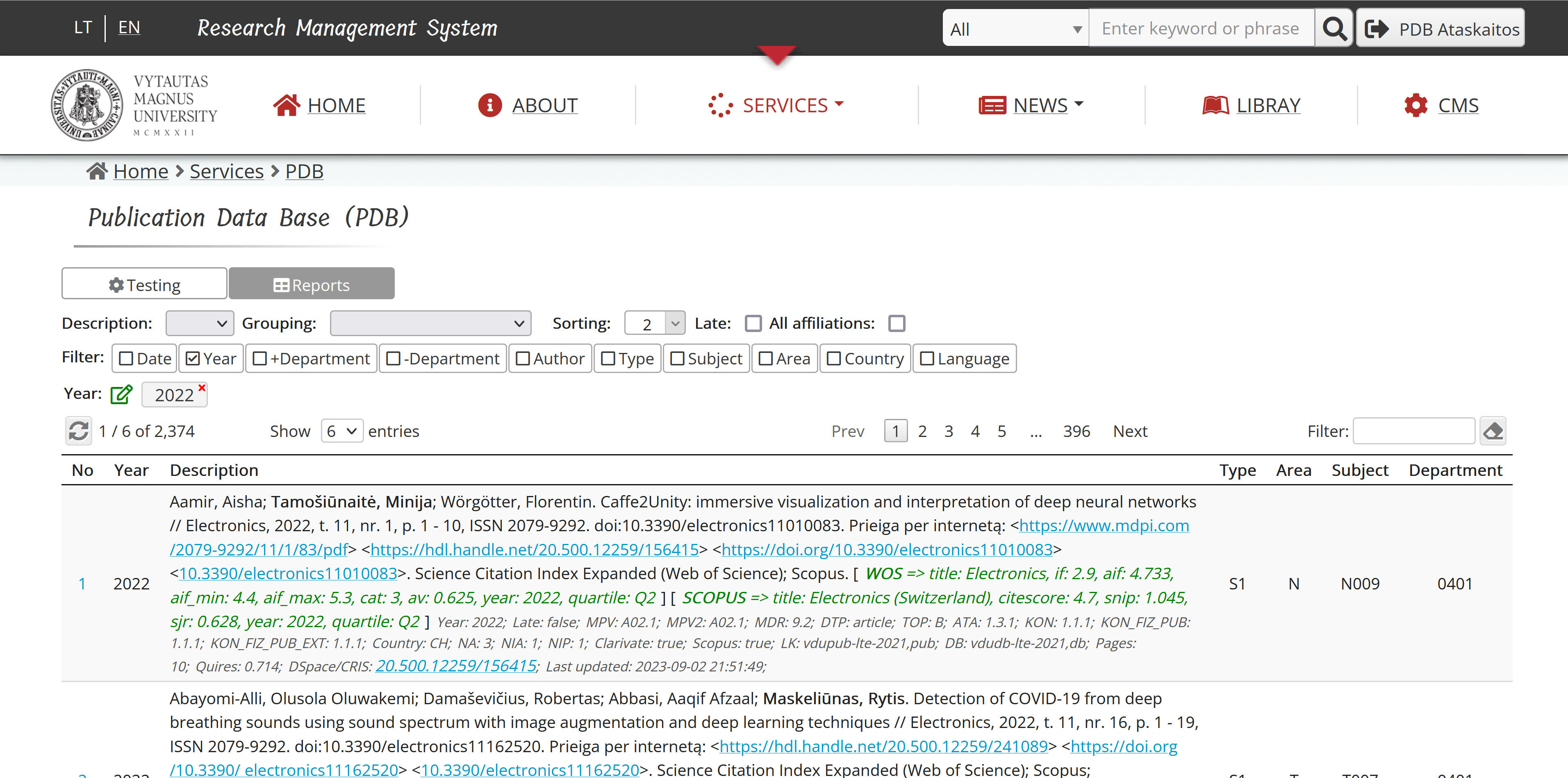
The user interface functions as an application in a web browser and is created using HTML, SCSS and jQuery software framework. This application communicates with the server application, which is developed in the PHP programming language. PostgreSQL RDBMS is used to store data. The realization of the system is based on KUSoftas CMS, which is used as the programming framework of the application. Rest API is created using PHP Slim micro framework, functionality of which is provided through KUSoftas CMS. With the help of this micro framework, additional request processing functions are developed, which, together with the main function of request processing, ensure the invariant output of reports in the following formats:
- HTML
- Word
- CSV (Excel)
Filtering, grouping, and sorting capabilities for the reporting request engine response:
- Year - filtering by the exact set of years.
- Date - filtering by the date range.
- +Department - filtering by affiliation department(s).
- -Department - elimination by affiliation department(s).
- Author - filtering by author(s).
- Type - filtering by type(s) of the publication.
- Research field - filtering according to research field(s) of the publication.
- Area - filtering by the research area(s) of the publication.
- Country - filtering by country(s) of the publication.
- Language - filtering by language(s) of the publication.
- Delayed - to include late (by reporting period) publications.
- All affiliations - to include such publications, which have no institutional affiliation, but is the author of the institution.
- Description - citation style.
- Grouping - grouping by departments and / or research fields.
- Sorting - the choice of sorting options.
The user interface has the possibility to test requests based on selected filtering possibilities. For this, the jQuery DataTables plugin is used, with the help of which a smart table is created to present the results of the request.
Top4. Advantages of Research Management System
| Open system architecture | Access to all system functions through REST API creates favorable conditions for integration with other University systems |
| Standards and recommendations: CERIF, COAR, OpenAIRE, etc. | Compatibility with essential standards and recommendations ensures efficient data exchange and import / export with other similar systems |
| Live metadata imports during submit: WOS, SCOPUS, PubMED, CrosssRef, eLABa, etc. | Makes it possible to quickly and efficiently prepare and submit research entities in the cataloguing process by importing records metadata from external live sources |
| Integration of researcher's profile with ORCID | Allows CRIS researchers to log-in to ORCID via CRIS and freely move their CRIS profile information to their ORCID profile and vice versa. |
| The content can be integrated as an integral part of the content of the University website | Creates conditions for the effective dissemination and evaluation of information in external evaluation systems on the basis of the Web by providing URL links to CRIS research entities as URL links to the University website – unambiguously identifying that this is scientific output of the University |
System configuration wide capabilities:
|
Creates opportunities for a very effective adaptation of the system to meet the needs of the University, in many cases avoiding complex programming work and enables to achieve results in a relatively short time |
| Open source code, advanced architecture and modern technologies | Base platform used by CRIS is rapidly developing - it is a promising solution for the future |
